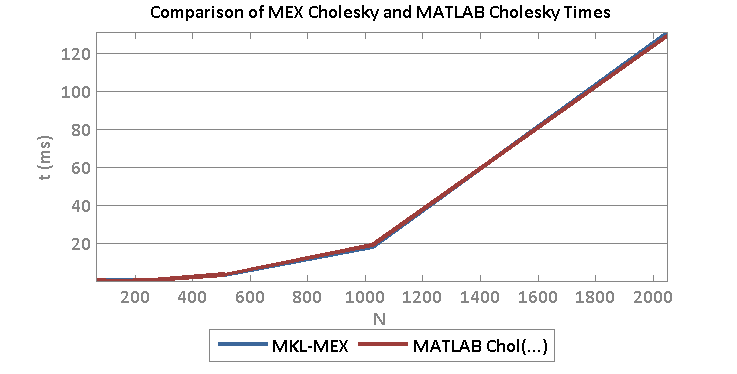
Matlab has incorporated GPU processing on the parallel computing toolbox and you can create GPU array objects using the gpuArray(…) function in MATLAB. I created a brief script to compare matrix multiply of a 2048 x 2048 matrix against a vector. Ordinarily, the CPU operations see reasonable speedup (~2x) from moving from double to single precision values. However, moving to the GPU implementation results in a speedup of 6.8x for Double and 5.6x for Single! This means that if you can take a matrix-vector multiply that is double precision and convert it to single precision GPU version, you may see a gain of nearly 14x.
The following we generated in Matlab R2014b on an i7-4770 3.5 Ghz CPU (8 CPUs) with 16GB Ram and a Geforce GTX 750.

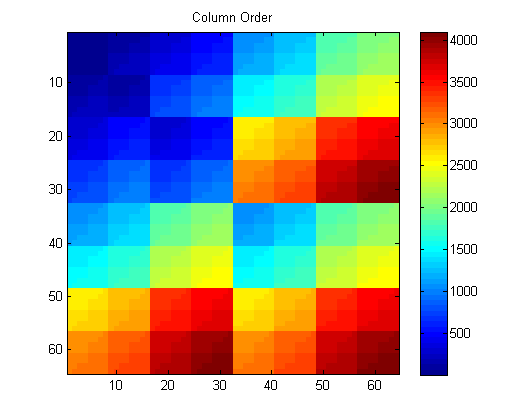
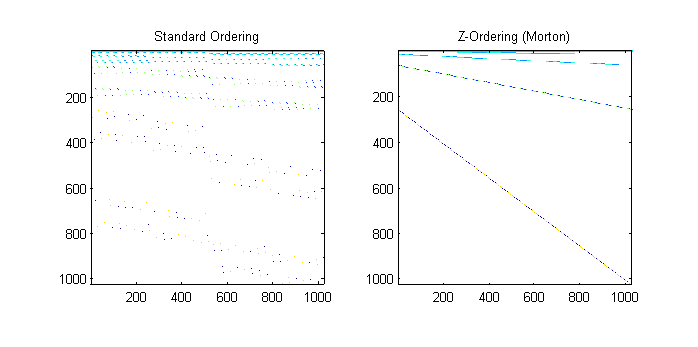
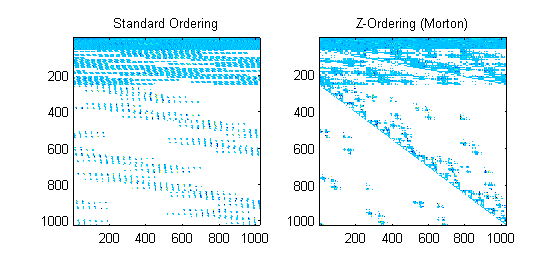
 The next step is to evaluate speed of the gpuArray on a basic L1 Optimization set—l1 magic.
The next step is to evaluate speed of the gpuArray on a basic L1 Optimization set—l1 magic.
The code used to generate this data is as follows:
allTypes = {'Double', 'gpuArrayDouble', 'Single', 'gpuArraySingle'};
allTimes = nan(length(allTypes),1);
n = 2048; % size of operation for Ax
num_mc = 2^10; % number monte carlo runs to compute time average of runs
randn('seed', 1982);
Am = randn(n,n);
xm = randn(n,1);
for ind_type = 1:length(allTypes)
myType = allTypes{ind_type};
switch lower(myType)
case 'double'
A = double(Am);
x = double(xm);
case 'single'
A = single(Am);
x = single(xm);
case 'gpuarraydouble'
A = gpuArray(Am);
x = gpuArray(xm);
case 'gpuarraysingle'
A = gpuArray(single(Am));
x = gpuArray(single(xm));
otherwise
error('Unknown type');
end
tic
for ind_mc = 1:num_mc
y = A*x;
end
allTimes(ind_type) = toc/num_mc;
end
%% Display the results
figure(34);
clf;
bar(allTimes*1000);
set(gca, 'xticklabel', allTypes, 'color', [1 1 1]*.97);
title(['Timing of CPU and GPU in M' version('-release')]);
xlabel('Type');
ylabel('Time (ms)');
grid on
%%
figure(35);
clf;
speedupLabels = {('Double to Single') , ...
('Double to gpuDouble'), ...
('Single to gpuSingle'), ...
'Double to gpuSingle'};
bar([allTimes(1)/allTimes(3), allTimes(1)/allTimes(2), ...
allTimes(3)/allTimes(4), allTimes(1)/allTimes(4)]);
set(gca, 'xticklabelrotation', 15, 'xticklabel', speedupLabels, 'color', [1 1 1]*.97);
title(['Speedup of CPU and GPU in M' version('-release')]);
xlabel('Type');
ylabel('Speedup');
grid on