This article will attempt to establish a performance baseline for creating a custom Cholesky decomposition in MATLAB through the use of MEX and the Intel Math Kernel Library (MKL). In the past I showed a basic and block Cholesky decomposition to find the upper triangular decomposition of a Hermitian matrix A such that A = L’L. The block performed substantially better than a single term operation but still not as good as MATLAB R2013a Intrinsic Chol(…) function. The times worked out to be that the non-blocking Cholesky decomposition in MATLAB was about 200x slower than MATLAB and the Blocking was about 7x slower. Further work by Chris Turnes in porting his code to C++ showed substantial gains, and using BLAS routines rather than naive C++ improved things further. However, parity with MATLAB was still not achieved. Sources indicate that MATLAB uses the MKL internally, so I correctly expected that calling the MKL LAPACK Chol(…)-equivalent function should yield equivalent results. However, the operation is blazing fast and so care must be taken with memory management and copies in order to reach parity with MATLAB.
The Intel MKL is a fast mathematics library from Intel that includes full implementations of BLAS, LAPACK, FFT, and other operations. It widely utilizes clever optimizations and Single Instruction Multiple Data (SIMD) operations like SSE. The LAPACK routine we are interested in is
int dpotrf(char *uplo, int *n, double *a, int*lda, int *info);
where dpotrf(…) is the double precision (“d”), symmetric or Hermitian positive definitite (“po”) triangular factorization (“trf”) LAPACK routine. We wish to create an upper triangular factorization by setting uplo to “U” for upper, the dimension n and lda to the size of the matrix, and the data pointer a to a COPY of the matrix to factor because this factorization is going to happen in place.
It turns out the copy operation is best done but copying only the upper triangular part of the matrix A. A naive copy using
plhs[0] = mxDuplicateArray(A);
is a poor choice for two reasons. First, the function dpotrf(…) writes its results only in the upper triangular portion of result and leaves the rest of the matrix as is. The result is that you must zero out the lower triangular part in C++ or call triu(C) on the resulting triangular factorization in MATLAB. Doing a FULL copy and then calling triu(…) results in code that is about 15% slower than only copying the parts of A you need and then dropping triu(…). The proper code for extracting the upper part of a given matrix was written by Turnes as:
plhs[0] = mxCreateDoubleMatrix(n, n, mxREAL);
double *data = mxGetPr(plhs[0]), *prIn = mxGetPr(prhs[0]);
for (int i = 0; i < n; i++)
memcpy(data + i*n, prIn + i*n, sizeof(double)*(i+1));
Once this is done, the LAPACK routine can be executed. As it turns out, you can run either a full copy of MKL or plug into the MATLAB version by compiling with mex and the -lmwlapack option like so:
mex cholchol.cpp -lmwlapack
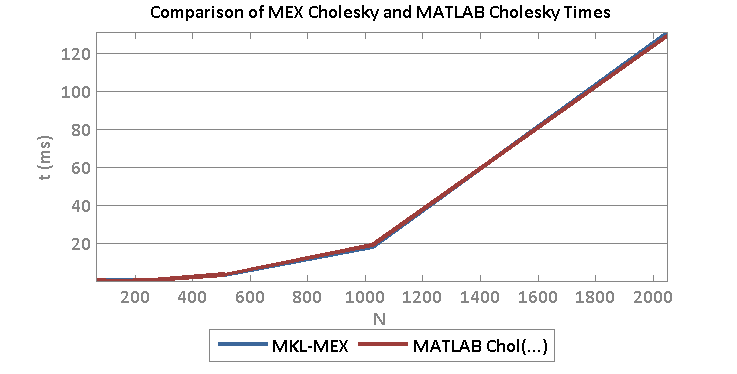
Both reach parity with MATLAB for large sizes. This means that we may be able to reach parity with MATLAB using MEX code provided that we use optimized BLAS routines. For example, at size 2048 x 2048 real matrix, I obtain the following timing:
- Time cholchol(…) MEX/MKL: 0.1315 s
- Time MATLAB: 0.1302 s
Next, I may consider implementing the block Cholesky decomposition in C++ using basic BLAS and the rank-k updates from MKL in order to attempt to achieve parity with more custom code.
You can download the main cholchol(…) code here.
And a comparison of times for various sizes:
This code as run on Sonata:
- Motherboard: Gigabyte X58U-UD3R
- Processor: Intel Core i7 950 @ 3.07 Ghz (8CPU)
- Memory: 8GB
- OS: Windows 7
- MATLAB: R2013A
- Compiler: Visual Studio 2012
- MKL: Intel Composer XE 2013 SP1